Here is a summary of the key points from the blog post:
title:: Detect mitotic figures in whole slide images with Amazon Rekognition
publisher:: AWS
people:: Pablo Nicolas Nuñez Pölcher, Razvan Ionasec
organization:: Amazon Web Services, University Hospital Essen, Technical University Munich
domain:: Machine Learning, Healthcare, Digital Pathology
Summary
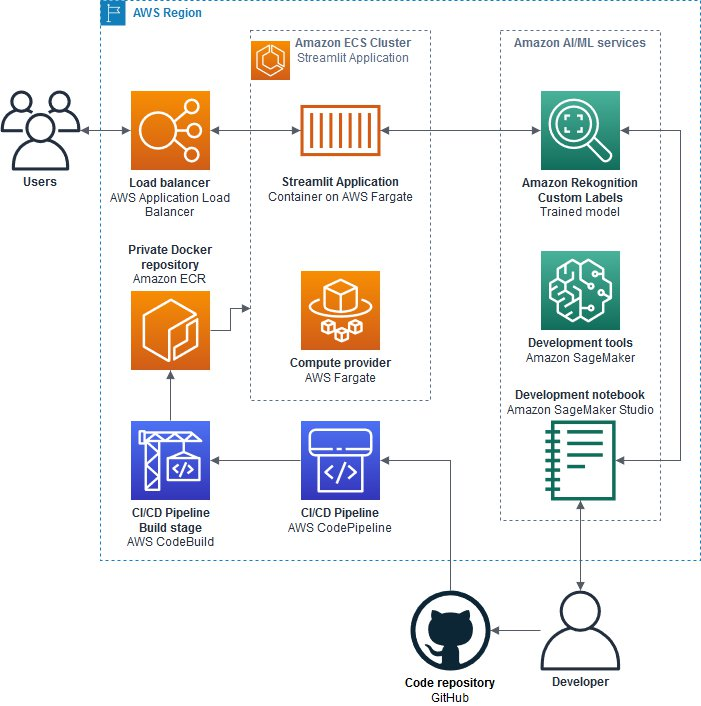
The blog post describes how to use Amazon Rekognition Custom Labels to train a model that detects mitotic figures in whole slide images of canine mammary carcinoma. The key steps include:
- Processing and sampling the whole slide image dataset using Amazon SageMaker Studio
- Training a custom object detection model using Amazon Rekognition Custom Labels
- Deploying a simple web application using Streamlit to demonstrate the model
- Setting up a CI/CD pipeline with AWS CodeBuild and CodePipeline to automate deployment
The solution allows developers without ML expertise to build an AI-powered digital pathology application using AWS services.
Data Points
- Uses dataset of canine mammary carcinoma whole slide images
- Processes images using Amazon SageMaker Studio and custom Python code
- Trains Amazon Rekognition Custom Labels model to detect mitotic figures
- Deploys demo Streamlit web app using AWS Fargate and Amazon ECS
- Sets up CI/CD pipeline with AWS CodeBuild and CodePipeline
- Estimated cost under $10 for running model and app for 1 hour
- Demonstrates how to integrate Amazon Rekognition into healthcare applications
- Authors thank Prof. Dr. Marc Aubreville for permission to use the dataset
- Highlights potential for ML to aid pathologists and accelerate diagnosis

